|
Gene prediction 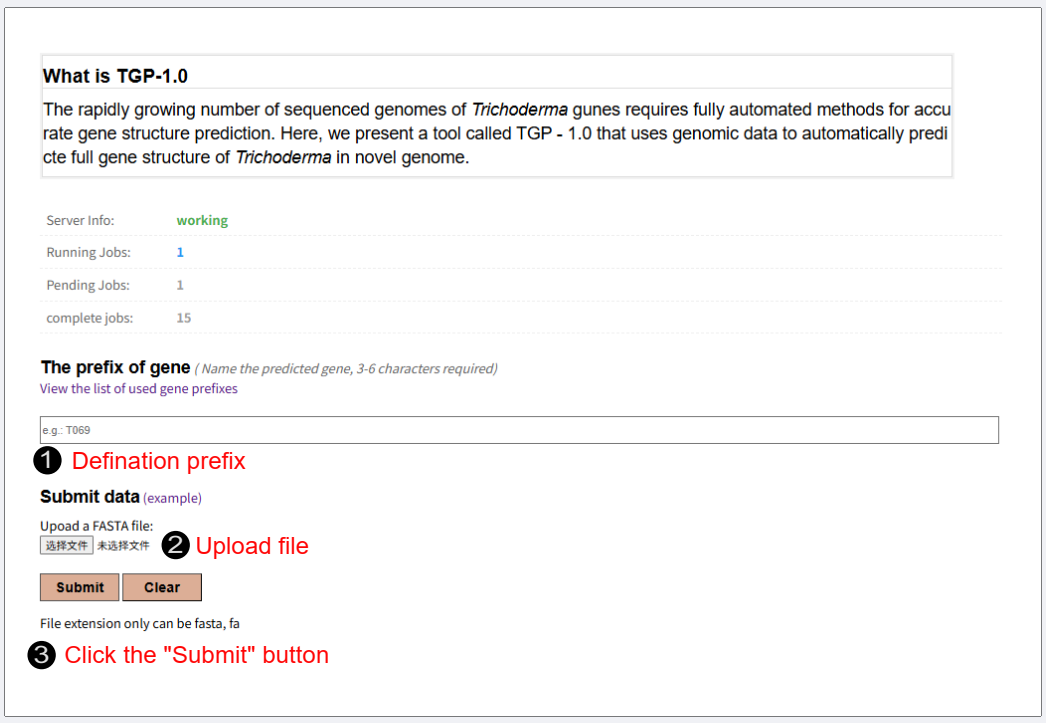
Prefix Please determine the gene prefixes that range from 3 to 6 characters as the identifier (ID) for gene prediction. (e.g., T069, FJ059) Gene prefixes cannot be repeated Upload file Please upload your genomic file in FASTA format (.fa, or .fasta extension). The maximum file size allowed is 100MB. Submit Click "Submit" button to queue your task. A tracking link will be generated, allowing you to monitor real-time progress (Pending, Running, Completed, or Failed) and download the results directly from this page. 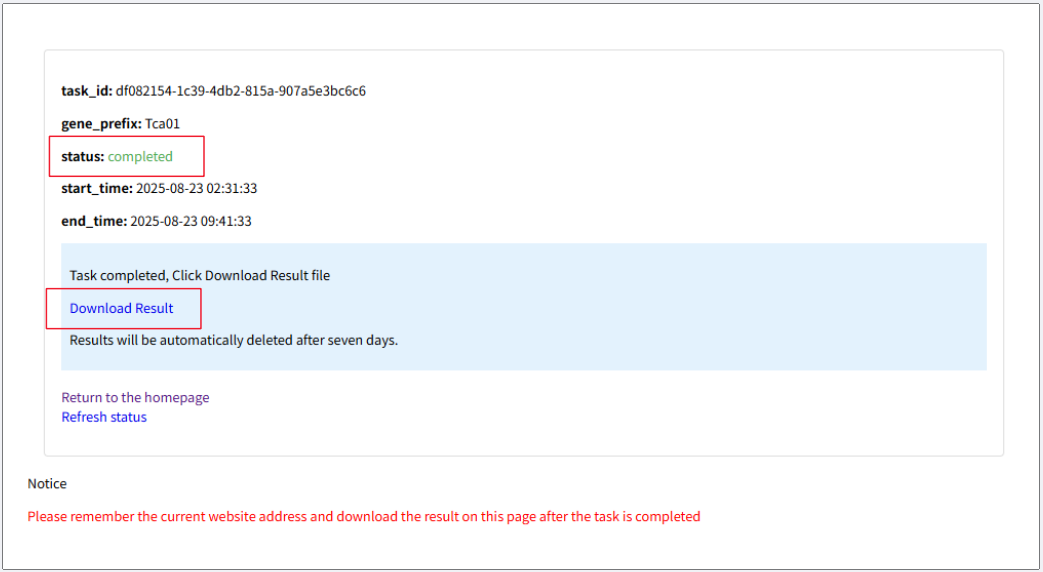
Download Result Upon completion (status: 'Completed'), click 'Download Result' to obtain a .zip folder containing three result files: predicted amino acid sequences (.pep), nucleotide sequences (.cds), and annotation information (.gff). Species 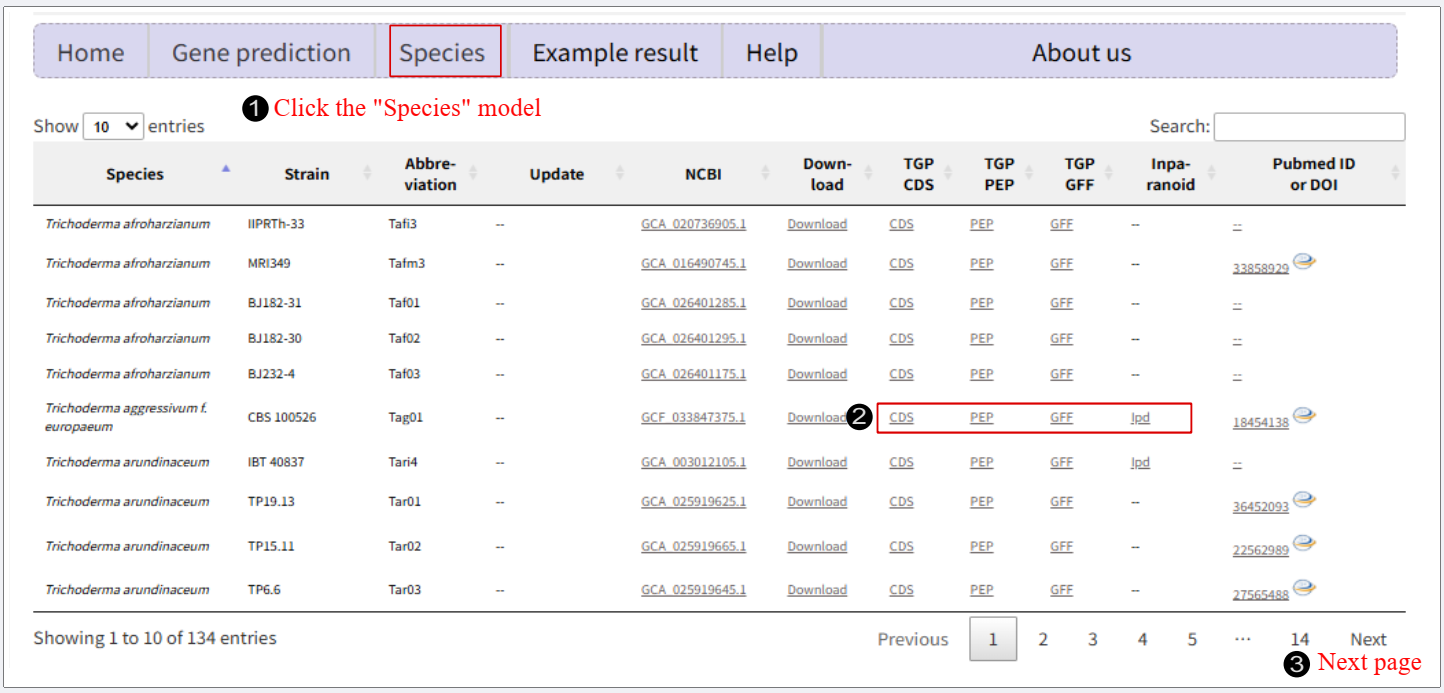
Species Click "Species" in the navigation box to enter the species module File of TGP prediction
Click 'CDS', 'PEP', or 'GFF' for the corresponding species to download: Additionally, we performed comparative analysis of predicted genes versus reporter genes using the InParanoid tool. Click 'ipd' to obtain relevant results. Notice: The TGP pipeline utilizes Augustus trained on Fusarium species. Owing to the close phylogenetic relationship between Fusarium and Hypocreales, this pipeline is also suitable for gene prediction in fungal species within the Hypocreales. |